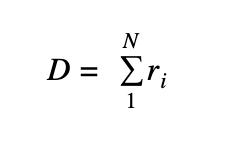Tutorial
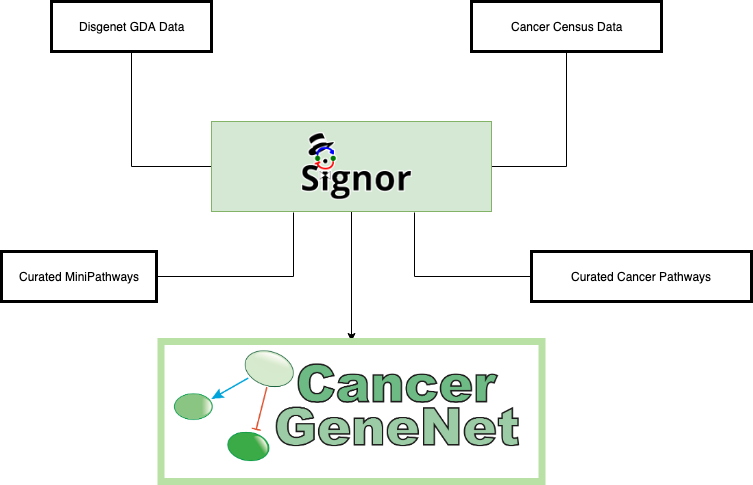
CancerGeneNET links links frequently mutated cancer genes to cancer phenotypes. The resource offers five options to browse its content:
-
Find Shortest Path to Cancer Hallmarks
-
Cancer Similarity Network
-
Cancer Browser
-
Connect Proteins
-
Curated Cancer Pathways
Find Shortest Path to Cancer Hallmarks
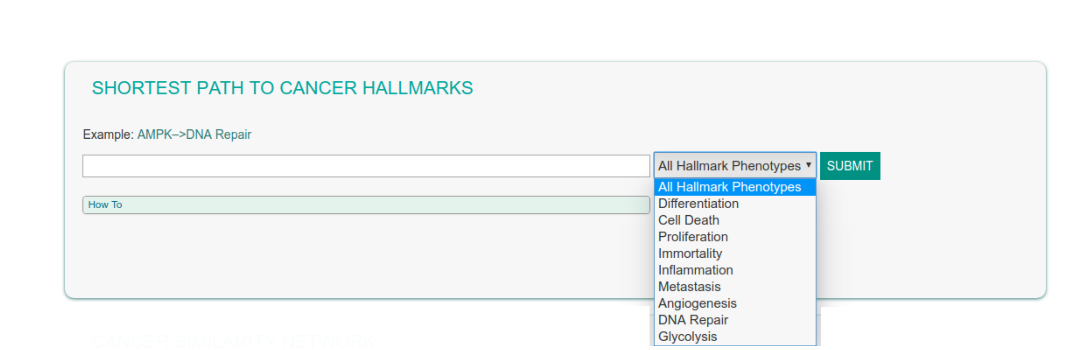
This functionality allows the user to look for network paths linking any gene to the cancer hallmark-phenotypes. Results are visualized as a graph and listed in a table.
In the Shortest Path to Cancer Hallmarks submit field, you can enter a gene or a list of gene names. By selecting a phenotype (or all the phenotypes) in a drop-down menu one obtains a graph showing the shortest path to the selected cancer hallmark.
Press the Submit button.
The query could require a matter of tens of seconds, as the graph algorithms explores a variety of paths in the network.
The result page displays a graph showing the shortest path(s) linking the selected gene/s to the selected cancer hallmark-phenotypes. To give to the user a wider perspective of the possible connections between the query gene and the selected phenotype(s) the paths that are one step longer than the shortest ones are also shown in the graph and displayed in the table.
The figure below shows the result page of a query looking for functional connections between the AMPK kinase complex and the DNA Repair phenotype.
-
Searched Proteins: AMPK
-
Selected Hallmark Phenotypes: DNA Repair
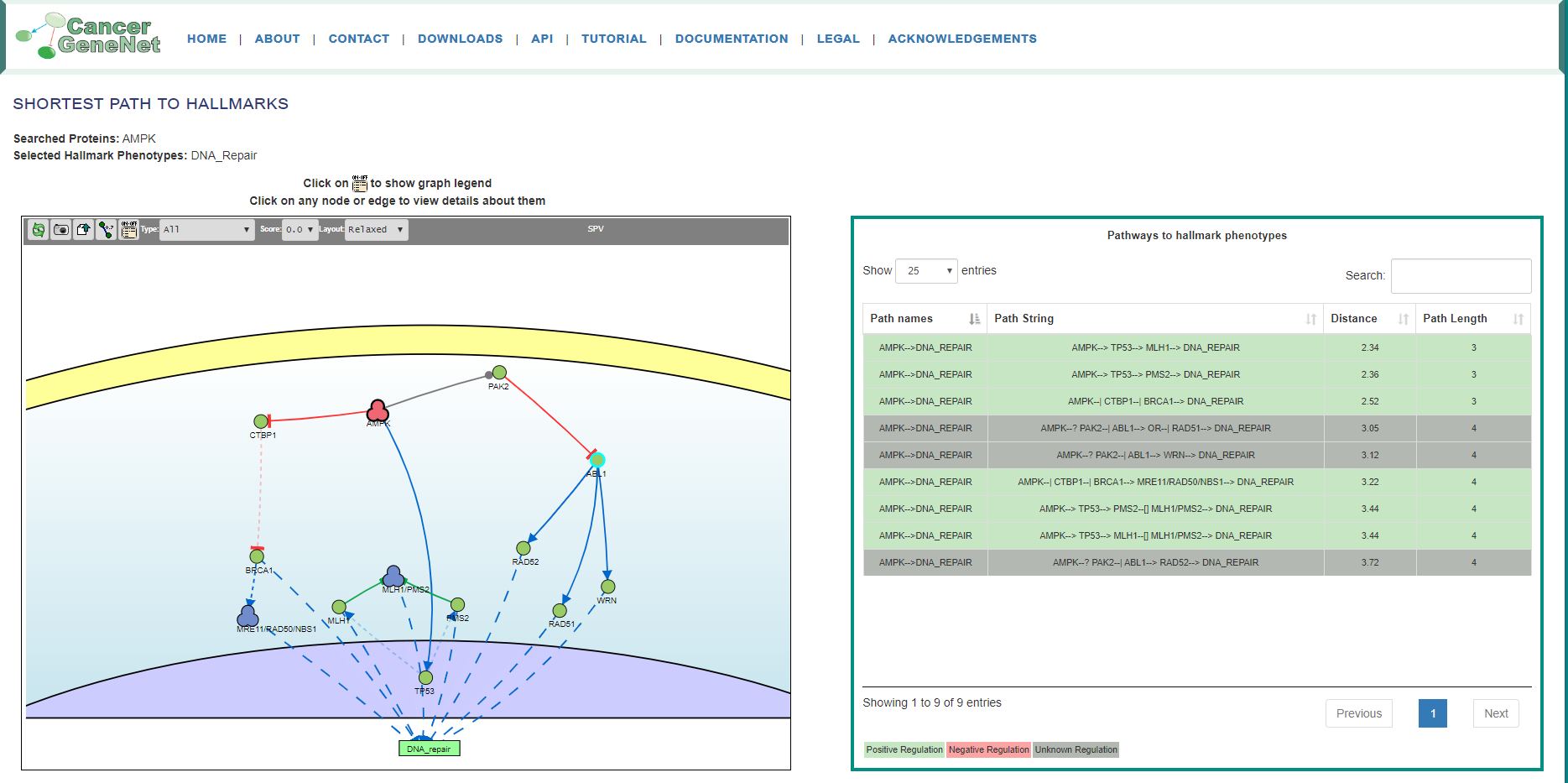
The query gene is highlighted in red in the graph and the resulting network displays all the connecting paths of minimal length. Each row of the summary table contains the following information path:
-
Path query: AMPK → DNA repair
-
Path string: each step is characterized by two nodes and an edge, the edge can have four different symbol according to type of relation effect.
-
→ activation
-
-| inhibition
-
-[] form complex
-
-? unknown effect
-
Distance: this parameter provides an estimation of the path length where the reliability of each step is taken into consideration. Each single-path step is associated to a reliability score r, taking into consideration a number of supporting evidence. The reliability score is converted into a distance by the following simpler relation:
d = 1-r
The final path score is the sum of each step distance:
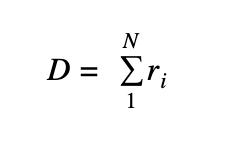
Where N is the number of steps in a path
-
Length: Number of steps in the path.
Depending on the even or odd number of inhibition steps, each path can be predicted to be activating or inhibiting for the target phenotype. This is represented in the table as a green or red background of the corresponding row. Whenever the path contains a relationship of unknown effect, the row is marked with a gray background. Finally, it is possible to download the annotation (with PMID reference) of the edges in each path as a tab-delimited file. The information in the Summary Table can also be downloaded. Each node and edge are labeled with different information, more details are provided in the following sections.
When the query is for 'All hallmark phenotype' the graph only shows the shortest path in order to limit the complexity of the resulting network display. However, the summary table contains all the retrieved paths grouped for each phenotype. The search box on the top right side allows to search a specific gene or phenotype of interest.

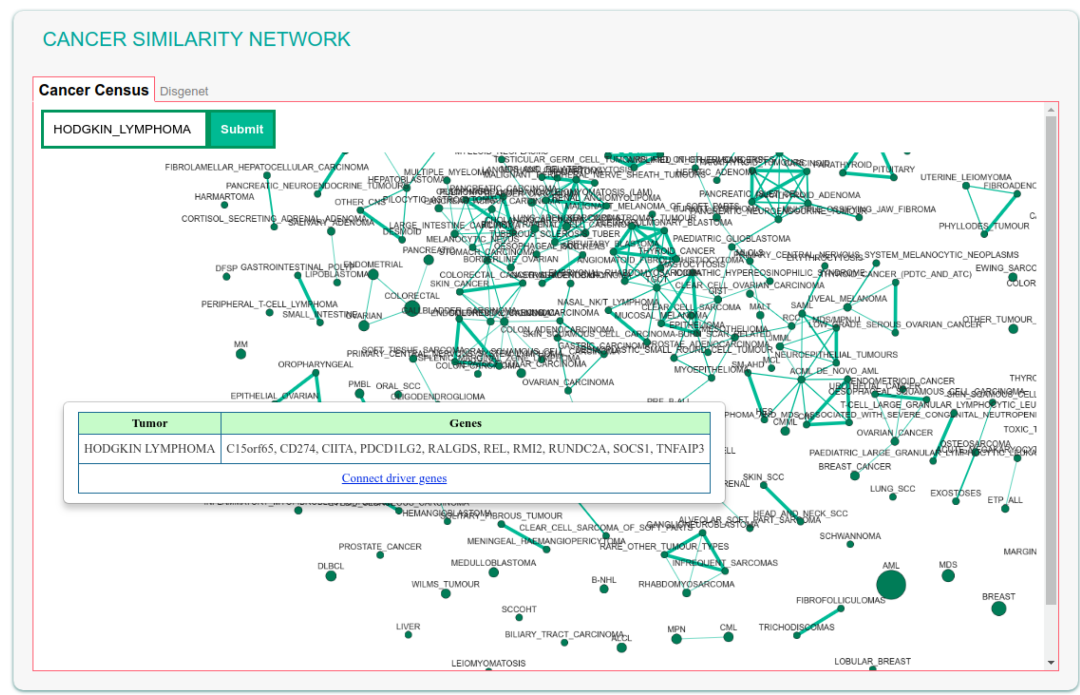
Cancer Similarity Network
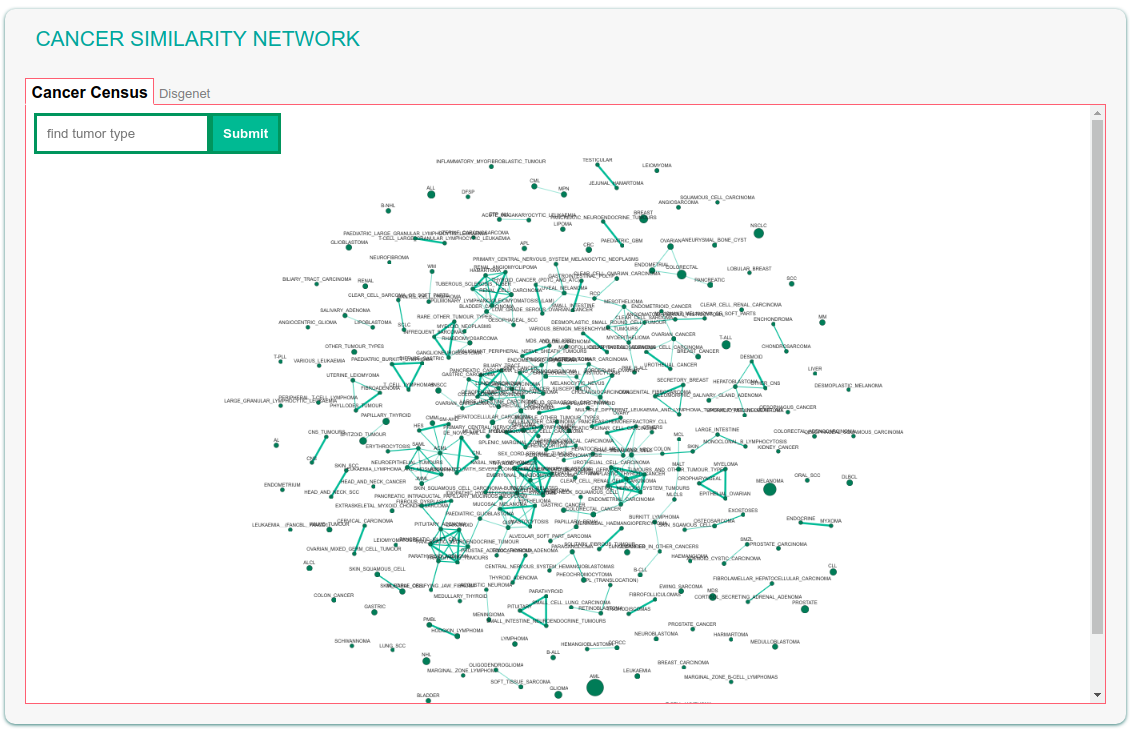
The Cancer Similarity Network offers a convenient entry point to the list of cancer genes annotated in the Cancer Gene Census of in DisGeNET. Each node in the graph is a tumor type. The node size is linked to the number of annotated cancer genes in each tumor.
The map also shows the connections between cancers that are most similar as evaluated by calculating the Jaccard index of the two tumor-specific lists of associated genes. Edge thickness is related to the value of the Jaccard index between the two linked tumors.
The graph can be explored by searching, via the “find tumor type” box, a specific tumor name, e.g.'Breast cancer'. The field also offers autocomplete functionality in order to facilitate searching by name. After submitting the query the corresponding node becomes larger and yellow for easy identification.
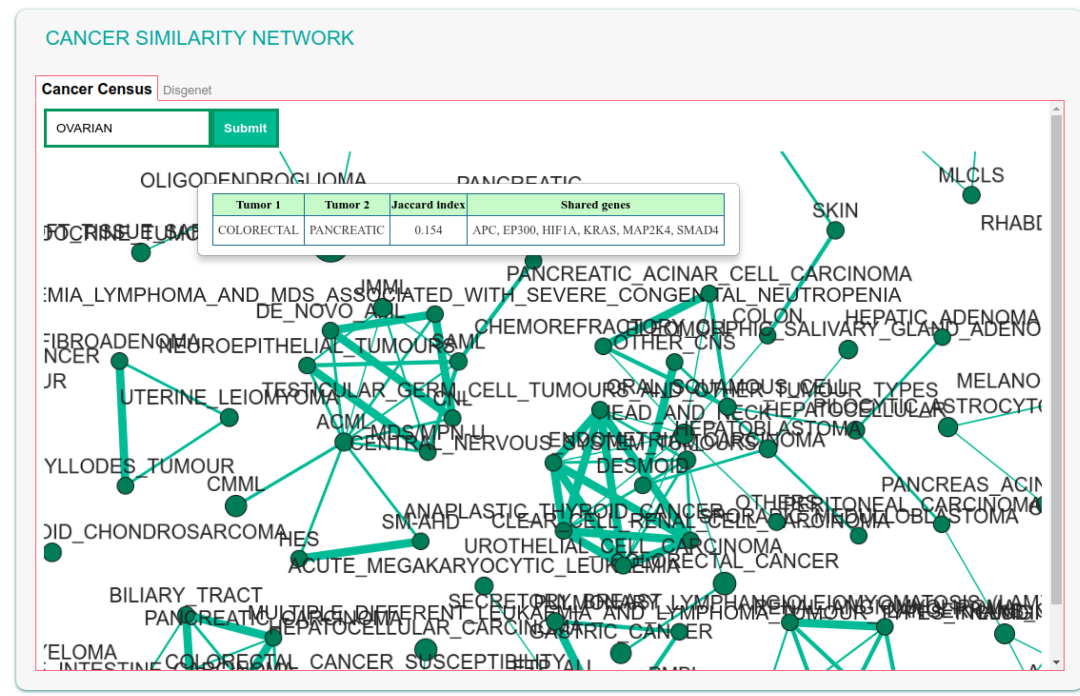
By clicking a node (cancer), a pop-up window appears showing the list of cancer associated genes and a hyperlink to the 'Connect driver genes' function. This function searches in the SIGNOR database for causal connections between the cancer genes in the list and displays a corresponding interactive graph.
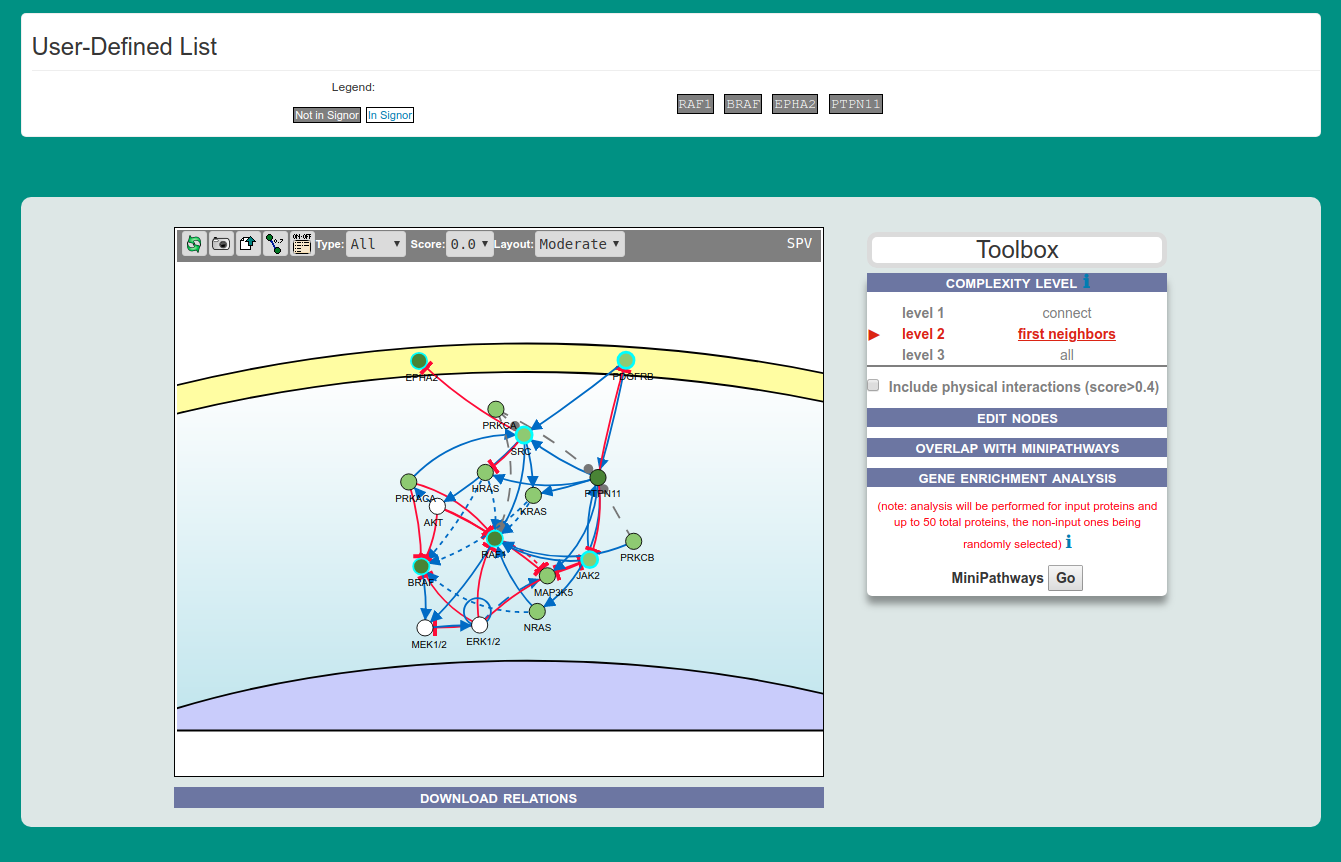
In the interactive viewer, nodes are proteins (or other entities considered in SIGNOR) and edges represent the causal relationships between them. The graphic viewer yields a dynamic, customizable display of the retrieved interactions, whose attributes are summarized with the use of symbols and color codes: direct interactions are displayed as solid lines and indirect ones as dashed lines, while edge color and arrow shape represent the effect (up- or down-regulation): up-regulations are represented as blue arrows, while down-regulations as ‘T-shaped’ red arrows. Nodes also have a color and shape code.
The query nodes (those in the cancer gene list) are illustrated as small, dark green circles, while first neighbours are light green. White circles and blue clover leaves represent protein families (e.g. ERK for MAPK1 and MAPK3) and complexes, respectively. Yellow squares represent small molecules and chemicals, while phenotypes and stimuli are rectangles.
Proteins, small molecules, stimuli, phenotypes and other entities are spatially organized in four main cellular compartments: the extracellular space, the plasma membrane, the cytoplasm and the nucleus. Nodes can be moved by the user to obtain a customizable layout of the entities. Every relationship is linked to a score.
Nodes that are targeted by specific anti-cancer drugs are highlighted by a cyan border.
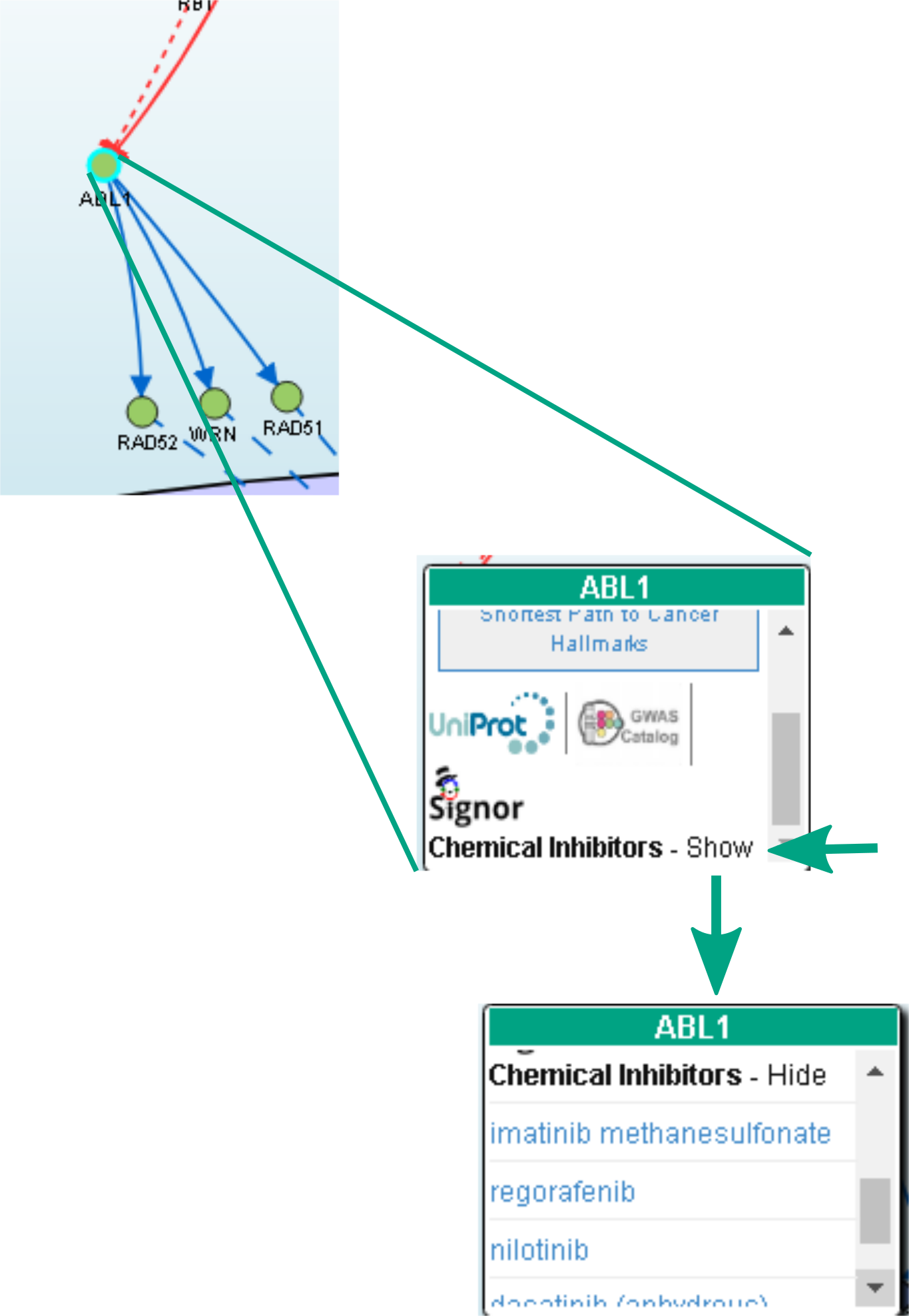
After clicking on a node, a pop-up window displays the entity details: name, external links (e.g. Uniprot, ChEBI, GWAS catalog), anti-cancer drugs associated to the node, the link to the related SIGNOR page and a link to calculate the Shortest Path to Cancer Hallmarks for the selected node.

By clicking on each
edge it is possible to obtain details about the interaction: the mechanism (e.g. phosphorylation, binding...), the cell line or organism in which the interaction has been observed, the literature reference and the sentence supporting the interaction.
By clicking on
symbols on the top bar of the visualizer it is possible:
-
Reset the layout
-
Take a screenshot of the graph
-
Export the network: download data underlying the graph representation
-
Show or hide the interaction score
-
Show or hide the graph legend
-
Filter the graph for interaction type and effect (up- down-regulate)
-
Select an edge score threshold
-
Select a more compact or “relaxed” layout where nodes are close or far apart
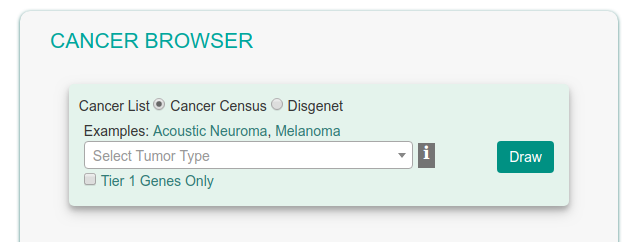
Cancer Browser
The Cancer Browser section offers an alternative way to access data and tools in the resource.
A drop-down menu allows the user to select a cancer type either from Cancer Census or from DisGeNET cancer lists. Clicking the ‘Draw’ button the user is lead to the results page showing an interactive graph viewer, as described in the 'Cancer Similarity Network' section. The graph viewer will display a schematic and detail-rich representation of the causal interactions between the cancer associated genes.
When exploring the Cancer Gene Census dataset, it is possible to consider only the most reliable associations (Tier 1) by checking the corresponding check boxes.
Connect Proteins
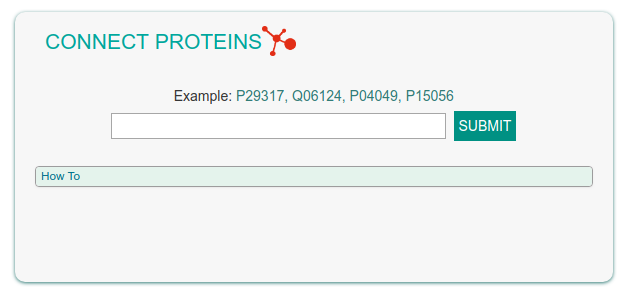
This functionality allows the user to enter a list of gene names (e.g. EPHA2, PTPN11, RAF1, BRAF) or UniprotKB IDs (e.g. P29317, Q06124, P04049, P15056) to obtain a graph linking the query proteins via causal interactions (see figure below).
Curated Cancer Pathways
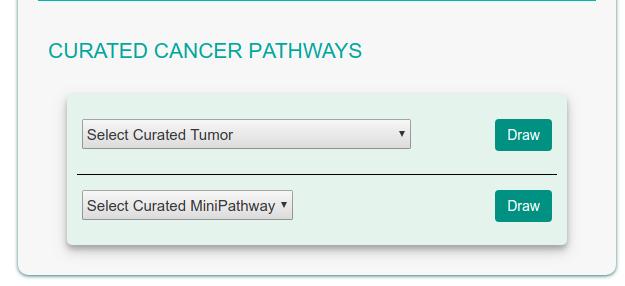
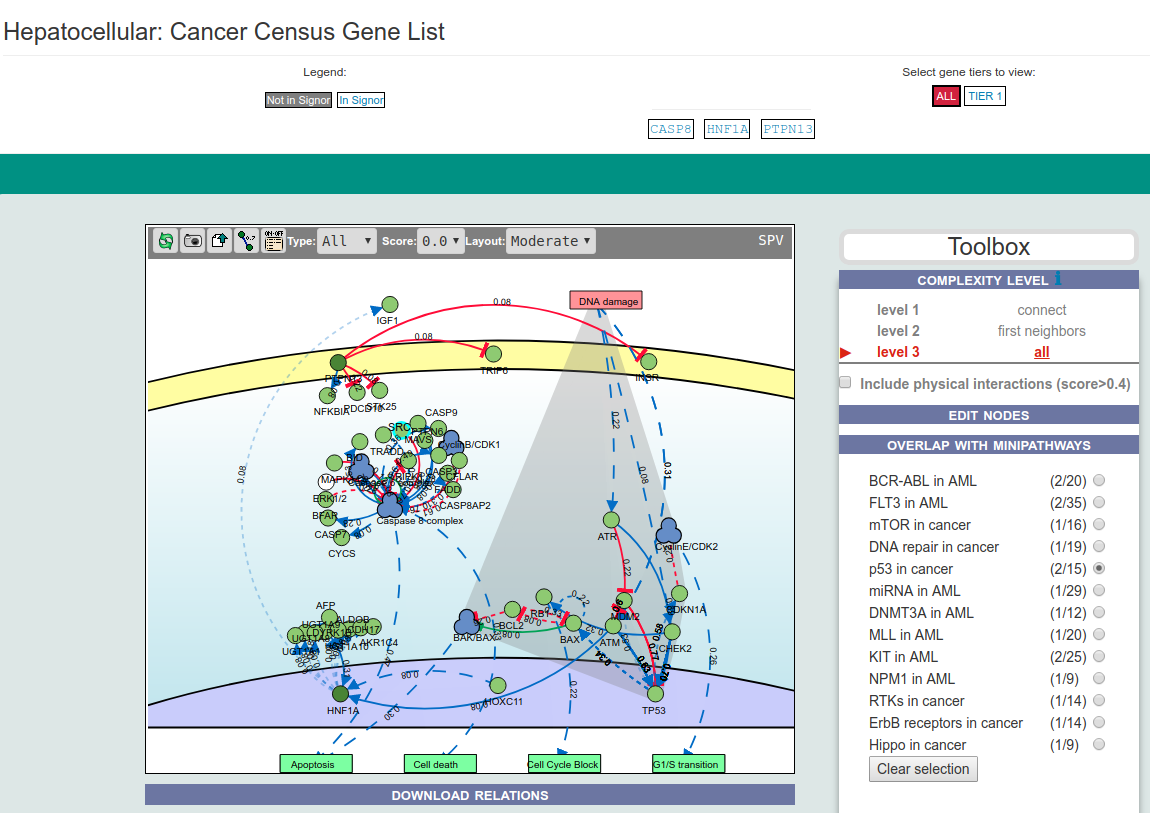
CancerGeneNet stores a set of curated cancer-specific pathways and Minipathways that are frequently mutated in different cancers. The graph illustrating the pathways can be displayed by selecting them in two drop-down menus in the bottom right frame of the homepage. The dropdown menus allows to select either a specific cancer pathway (e.g. Colorectal Carcinoma) or to select a mini pathway (e.g. ErbB Signaling). A MiniPathway is a small, manually curated pathway that is frequently altered in different tumors.
The pathway page shows an interactive display of the selected pathway.
Pathways are manually curated and the number of entities considered in each pathway is arbitrarily limited to the ones that are considered central, in order to have an interpretable graph. Each edge and node is clickable to get more information.
Entities are automatically laid out and assigned to specific cell compartments (extracellular space, membrane, cytoplasm, nucleus).
"Stimuli" and "Phenotypes" are always placed upstream and downstream respectively. A series of filters at the top of the graph frame allow the user to modify the display of the pathway by including or excluding indirect interactions or relationships below a score threshold.
It is possible to add (or remove) entities to the displayed graph by using the dedicated box on the right of the graph. This same toolbox also allows the user to perform additional analyses (see next section).
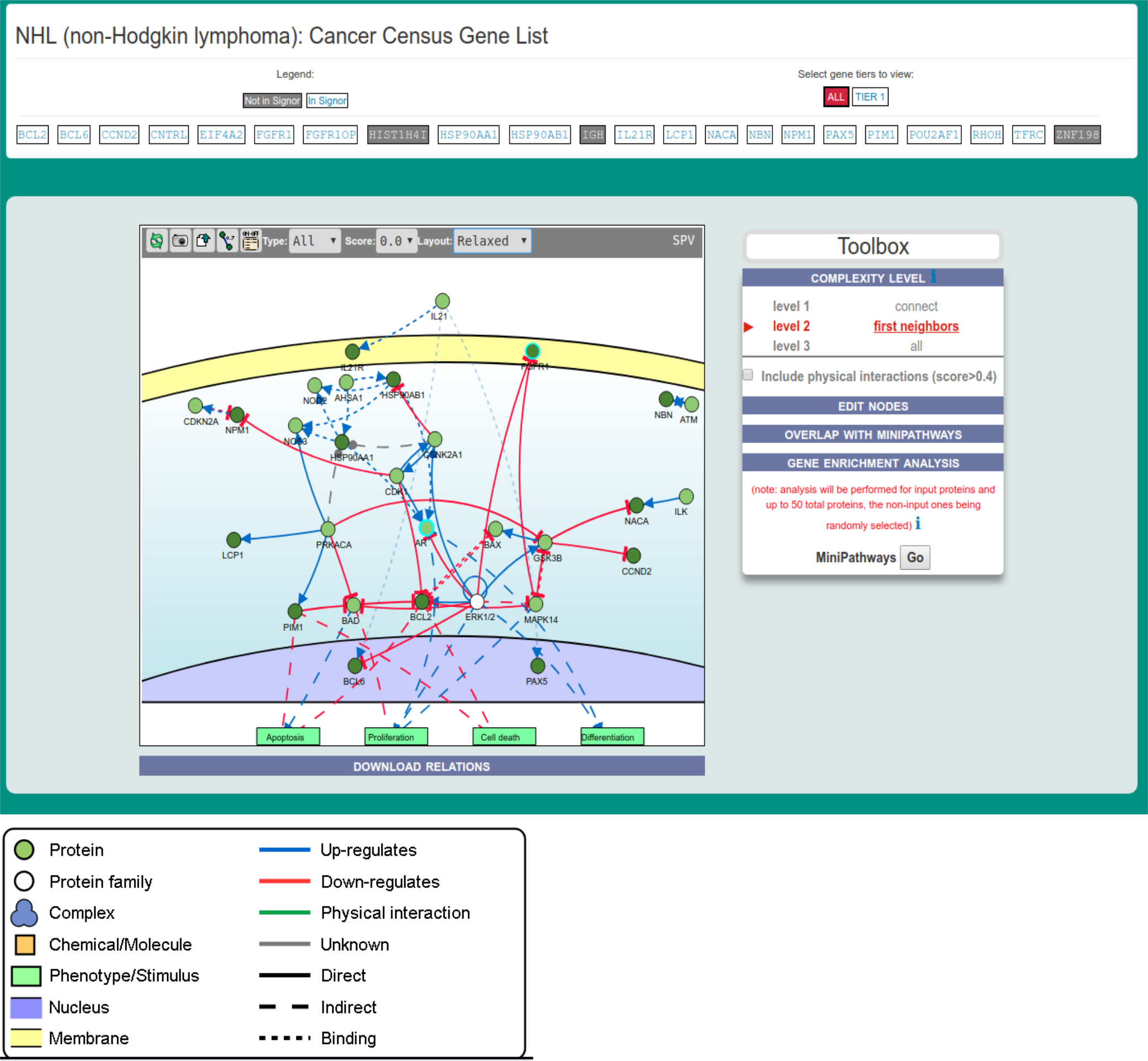
Toolbox functionality
The Toolbox section allows to perform different analysis:
-
Visualize the pathway at different levels of complexity
-
Edit Nodes
-
Overlap with MiniPathways
-
Gene set enrichment analysis
Visualizing the pathway at different levels of complexity
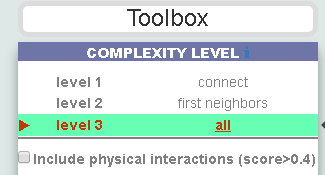
The box allows the user to display interactions at different levels of complexity using three different query strategies to retrieve interactions from the SIGNOR database. Connect (Level 1) searches for relationships connecting any two entities in the query list; First Neighbors (Level 2) is a multi-step strategy that initially performs a search in SIGNOR for all interactions involving any of the seed entities, next prunes nodes with degree-one leaving in the resulting network only the query proteins and 'bridge' proteins that help connecting them; All (Level 3) retrieves all signaling interactions involving any of the seed entities and the proteins in the SIGNOR database without any further filtering. By default the visualizer displays Level 2 interactions (First Neighbours); if no interaction is retrieved at level 2,level 3 is shown;
By clicking on Add physical interactions from Mentha the user can also include in the graph protein-protein interactions from the mentha database involving any seed entities and filtered by score. Only interactions whose reliability score is higher than 0.4 are shown. By clicking on Download Relations it is possible to download all the interaction visualized in the viewer (note: this does not include mentha interactions).
Edit nodes
The edit nodes functionality allows the user to edit the list of the seed entities, by removing or adding nodes.
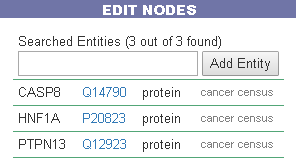
Overlap with cancer Mini pathway
This functionality shows the overlap between the query genes list and the gene list in the curated MiniPathways.
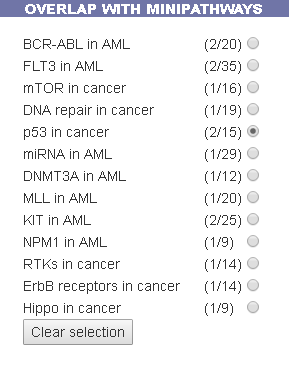
By selecting the radio button of a specific MiniPathway(eg. p53 in cancer), the MiniPathway is added to the interactive graph and the network area in common between the two graphs will be outlined with a red background.
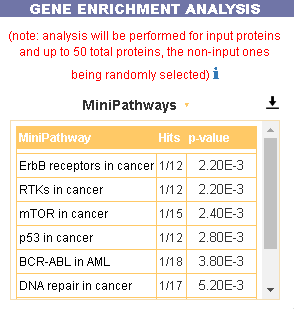
Gene set enrichment analysis
It is also possible to search whether the list of nodes in the displayed network is enriched for genes that are also annotated in MiniPathways. For this task go to the Gene Enrichment Analysis frame and click the GO button. The algorithm compares the list of genes that are displayed in the graph with the lists of pathway genes as annotated in the MiniPathways datasets.
The tool returns a table showing the MiniPathways that are most represented in the query cancer genes list. For each pathway, in the Hits column, the tool returns the ratio of shared genes. The p-value column provides the p-value calculated as follows:
-
We calculate the overlap between the gene list in a specific MiniPathway and the one in the displayed graph
-
We randomly select 10000 sets of the same size of the query gene list and calculate the overlap with the gene list of the specific MiniPathway
-
We count how many times we observe in the random set a match greater than or equal to the one obtained from the starting set and we divide this number by the total number of iterations
NOTE: to limit server overload, a maximum of 50 proteins will be used as input for each gene list in this analysis.
Download Page
Data can be downloaded in two different ways:
-
Through the Download page accessible via the top menu bar.
-
Through the interactive map by clicking on export on the top toolbar.
In the download page it is possible to download the CancerGeneNet datasets: